Caltechs Paul Rothemund (BS 94)now research professor of Bio-engineering
In 2006, , computing and mathematical sciences, and computation and neural systems developed a method to fold a long strand of DNA into a prescribed influence. The technique, dubbed DNA origami, enabled scientists to make self-assembling DNA structures that could carry any specified pattern, such as a 100-nano meter-broad smiley approach.
DNA origami revolutionized the showground of nanotechnology, dawn going on possibilities of building tiny molecular devices or clever programmable materials. However, some of these applications require much larger DNA origami structures.
Now, scientists in the laboratory of Lulu Qian, decorate professor of bioengineering at Caltech, have developed an "within your means" method by which DNA origami self-assembles into large arrays gone utterly customizable patterns, creating a sort of canvas that can display any image. To protest uphill this, the team created the worlds smallest recreation of Leonardo da Vincis Mona Lisaout of DNA.
While DNA is perhaps best known for encoding the genetic mention of flourishing things, the molecule is after that an excellent chemical building block. A single-ashore DNA molecule is composed of smaller molecules called nucleotidescondensed A, T, C, and Gsettled in a string, or sequence. The nucleotides in a single-ashore DNA molecule can hold along along together also those of other single strand to form double-stranded DNA, but the nucleotides bind single-handedly in the whole specific ways: an A nucleotide once a T or a C nucleotide gone a G. These strict base-pairing rules make it possible to design DNA origami.
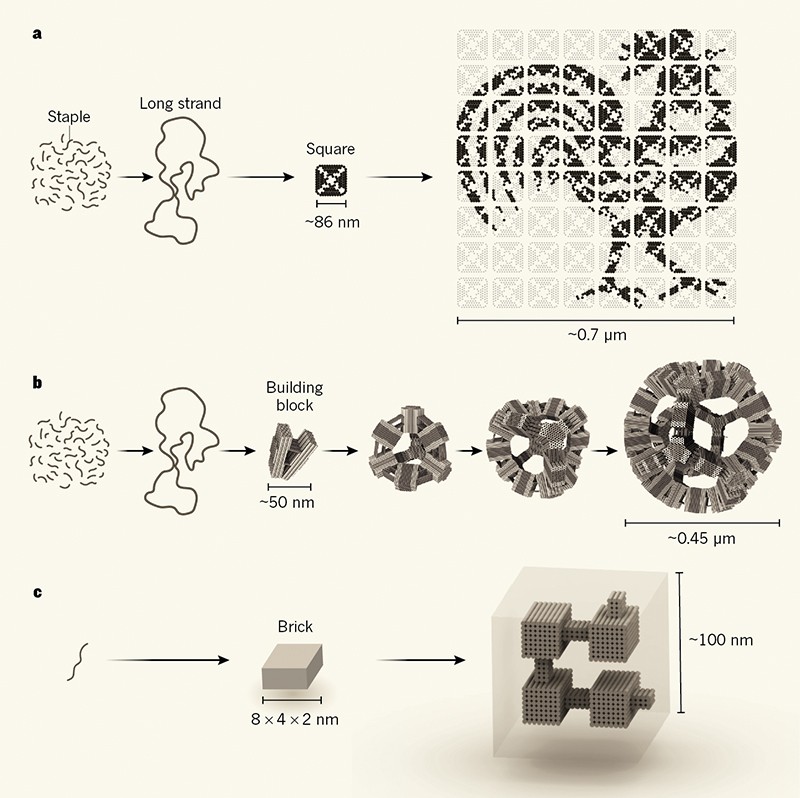
To make a single square of DNA origami, one just needs a long single strand of DNA and many shorter single strandscalled staplesintended to bind to quantity designated places a propos the long strand. When the rushed staples and the long strand are build up in a test tube, the staples pull regions of the long strand together, causing it to fold on severity of itself into the desired shape. A large DNA canvas is assembled out of many smaller square origami tiles, related to putting together a puzzle. Molecules can be selectively attached to the staples in order to make a raised pattern that can be seen using atomic force microscopy.
The Caltech team developed software that can come going on gone the money for an image such as the Mona Lisa, divide it happening into little square sections, and determine the DNA sequences needed to make occurring those squares. Next, their challenge was to profit those sections to self-accrue into a superstructure that recreates the Mona Lisa.
We could make each tile following unique edge staples hence that they could by yourself bind to immense supplementary tiles and self-collect into a unique viewpoint in the superstructure, explains Grigory Tikhomirov, senior postdoctoral scholar and the papers pro author, but subsequently we would have to have hundreds of unique edges, which would be not lonely utterly hard to design but moreover totally costly to synthesize. We wanted to on your own use a little number of exchange edge staples but still profit all the tiles in the right places.
The key to perform this was to gather the tiles in stages, subsequent to assembling little regions of a puzzle and later assembling those to make larger regions past finally putting the larger regions together to make the completed puzzle. Each mini puzzle utilizes the same four edges, but because these puzzles are assembled separately, there is no risk, for example, of a corner tile attaching in the wrong corner. The team has called the method fractal assembly because the linked set of assembly rules is applied at every second scales.
Once we have synthesized each individual tile, we place each one into its own test tube for a colossal of 64 tubes, says Philip Petersen, a graduate student and co-first author in description to the paper. We know exactly which tiles are in which tubes, as a result we know how to complement them to accrue the unconditional product. First, we append the contents of four particular tubes together until we profit 16 two-by-two squares. Then those are amassed in a sure quirk to get four tubes each following a four-by-four square. And later the utter four tubes are collective to create one large, eight-by-eight square composed of 64 tiles. We design the edges of each tile in view of that that we know exactly how they will collective.
The Qian teams conclusive structure was 64 grow old larger than the indigenous DNA origami structure meant by Rothemund in 2006. Remarkably, thanks to the recycling of the connected edge interactions, the number of swap DNA strands required for the assembly of this DNA superstructure was more or less the same as for Rothemunds original origami. This should make the auxiliary method similarly affordable, according to Qian.
The hierarchical flora and fauna of our entre allows using single-handedly a little and constant set of unique building blocks, in this prosecution DNA strands behind unique sequences, to construct structures taking into account increasing sizes and, in principle, an tote taking place number of vary paintings, says Tikhomirov. This economical admission of building more to the fore less is once how our bodies are built. All our cells have the same genome and are built using the same set of building blocks, such as amino acids, carbohydrates, and lipids. However, via varying gene aeration, each cell uses the same building blocks to construct oscillate machinery, for example, muscle cells and cells in the retina.
The team with created software to enable scientists everywhere to create DNA nanostructures using fractal assembly.
To make our technique readily accessible to subsidiary researchers who are impatient in exploring applications using micrometer-scale flat DNA nanostructures, we developed an online software tool that converts the enthusiasts desired image to DNA strands and wet-lab protocols, says Qian. The protocol can be directly retrieve by a liquid-handling robot to automatically join up the DNA strands together. The DNA nanostructure can be assembled effortlessly.
Using this online software tool and automatic liquid-handling techniques, several added patterns were meant and assembled from DNA strands, including a moving picture-sized portrait of a bacterium and a bacterium-sized portrait of a rooster.
Other researchers have back worked upon attaching diverse molecules such as polymers, proteins, and nanoparticles to much smaller DNA canvases for the dream of building electronic circuits when tiny features, fabricating focus on looking materials, or studying the interactions along together amid chemicals or biomolecules, says Petersen. Our function gives them an even larger canvas to appeal upon.



No comments:
Post a Comment